

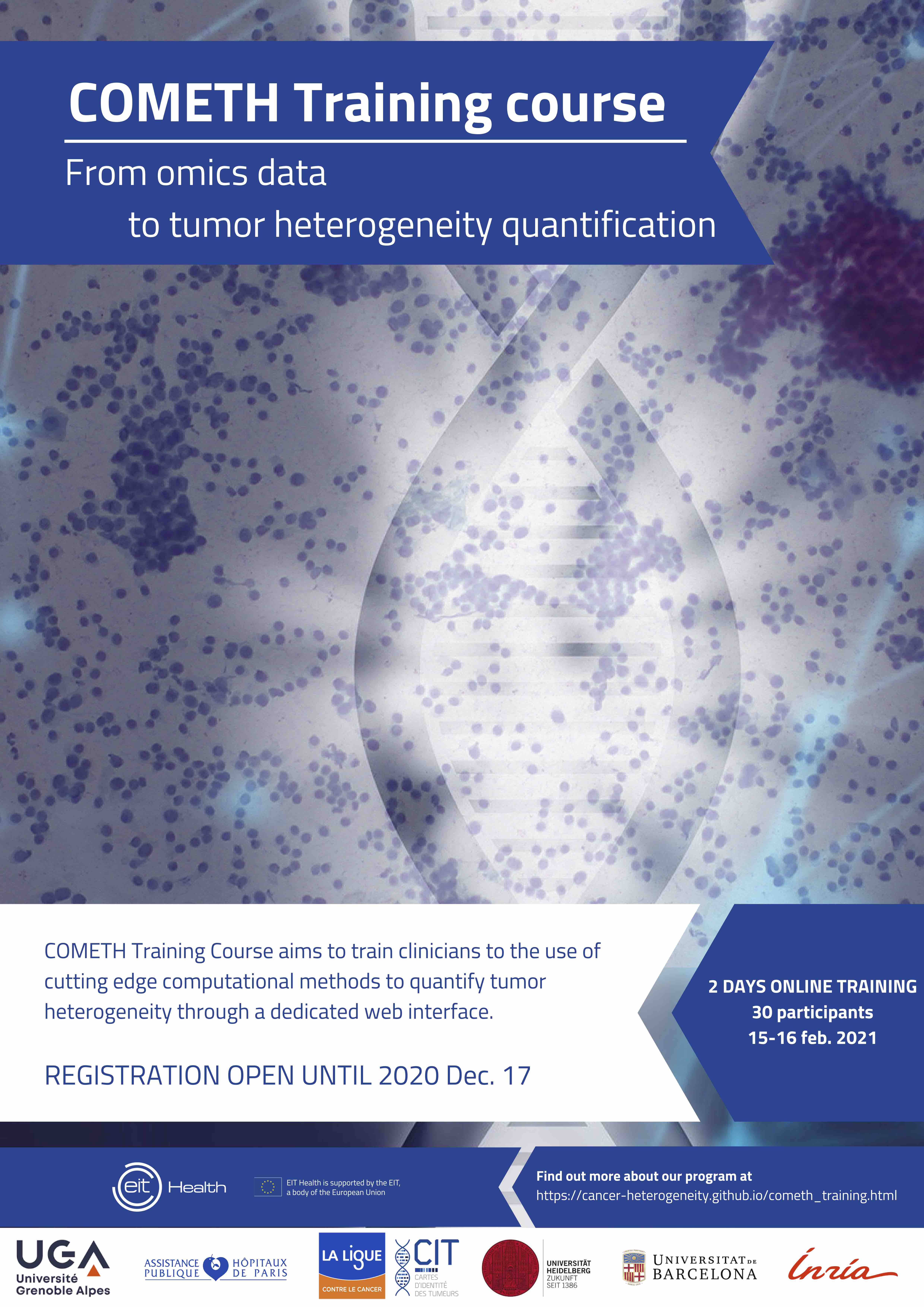
15-16 Feb.2021, ONLINE
This 2-day online course aims to train clinicians to the use of cutting edge computational methods to quantify tumor heterogeneity through a dedicated user-friendly web interface.
Registration deadline extension: 10 January 2021
For all participants:
For all participants:
For computational participants:
For clinician participants:
For those who want to run the deconvolution and interpretation app locally, we provide a docker image allowing to run all steps of the analysis locally, through your web browser. We provide the instructions for uses running a macOS or linux operating system!
cd ~
docker pull ashwinikrsharma/mrna_meth_decon:latest
git clone https://github.com/ashwini-kr-sharma/ShinyCompExplore.git
docker run --rm -it -p 3838:3838 -v ~/ShinyCompExplore:/srv/shiny-server/ ashwinikrsharma/mrna_meth_deconlocalhost:3838 in the address barGood to go!
Practical work will differ according to your affectation (medical or bioinformatic group). Please check your affectation and download the corresponding material here.
We have prepared toy expression datasets from the TCGA public cancer database (including clinical annotation tables).
!!Format!! .cvs with separator = “;” and decimal = “.” Samples in column, genes in row (use Gene Symbol as gene ID). If you want to use your own dataset during the course, please use the same csv format (separator = “;” and decimal = “.”).
Omic type : transcriptome
Normalized counts (linear scale)
FPKM (linear scale)
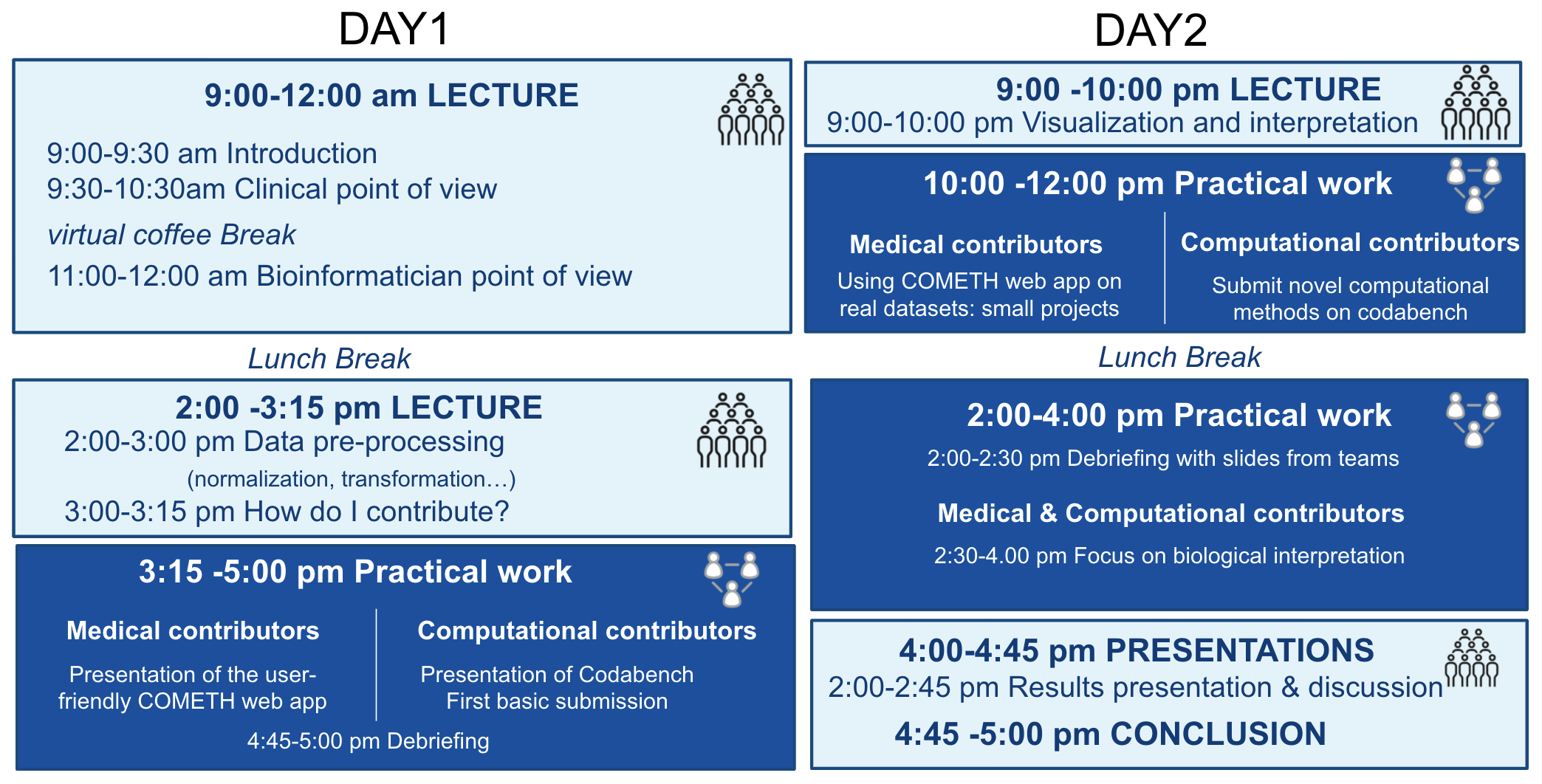
2-day online sessions with general lectures and practical training.
General lectures will cover:
• Introduction to cancer heterogeneity
• Introduction to computational methods
• Interpretation and visualization of the results
Cancer pathologists
Research scientists
Clinicians
Limited to 30 participants.
Registration is free of charge but mandatory.
Deadline: 10 January 2021
Technical support
Clémentine Decamps, Phd Student, Uni. Grenoble Alpes, France
Yasmina Kermezli, Postdoctoral fellow, Uni. Grenoble Alpes, France
